IMC12
In August, 2024 International Mycological Congress took place in Maastricht, Netherlands and some MMC members also attended. Professor Leho Tedersoo summarised his findings in a seminar that can be viewed here:
A short questionnaire with 7 questions about eDNA/sequence-based species description was shared among the participants of IMC12. We got responses from 21 people of whom 10 were PhD students, from the rest most were employed as researchers or professors and some chose not to reveal their academic positions. The average age for the respondents was 40 years old and most respondents are currently working in a research institute in EU. The respondents were asked to encircle suitable answers so each person could choose more than one option.
In question 1 the respondents were asked to specify what could be used as type-material for species description.
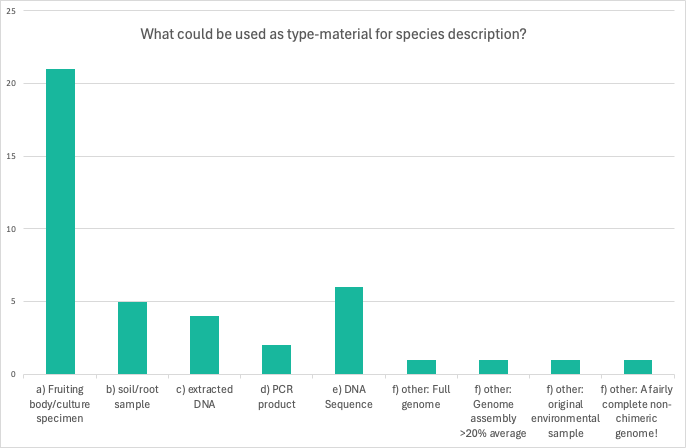
The most popular answer was that the best option is to use the fruiting body/culture specimen which everyone chose. There was also on one open-ended answer “Other” which 4 of the respondents chose. Their added options can be seen on the graph below.
Question 2: What should be the minimum requirement(s) for a sequence to be used in typification?
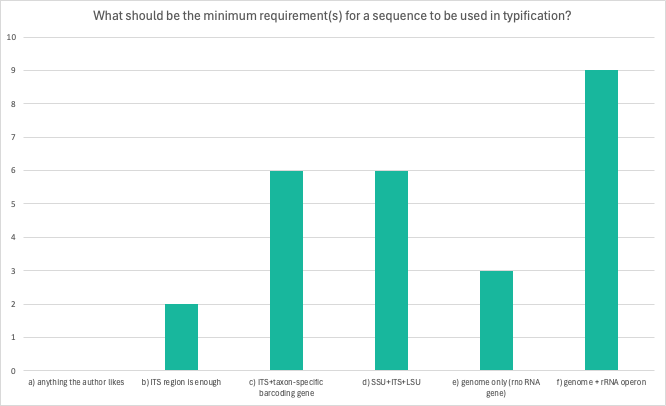
No clear understanding among researchers like with question 1. However, it is clear that “anything the author likes” is not an option. Some respondents added their ideas in the margins, e.g. one respondent added option g) presence of a physical specimen for a minimum requirement. One respondent wished to clarify that genome+rRNA operon is an option only with phylogeny.
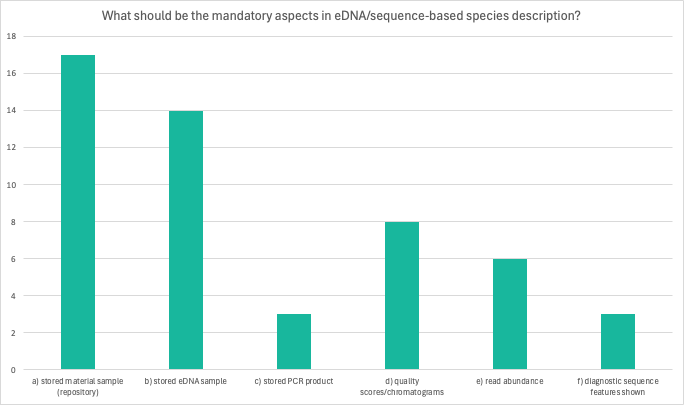
Question 3: What should be the mandatory aspects in eDNA/sequence-based species description?
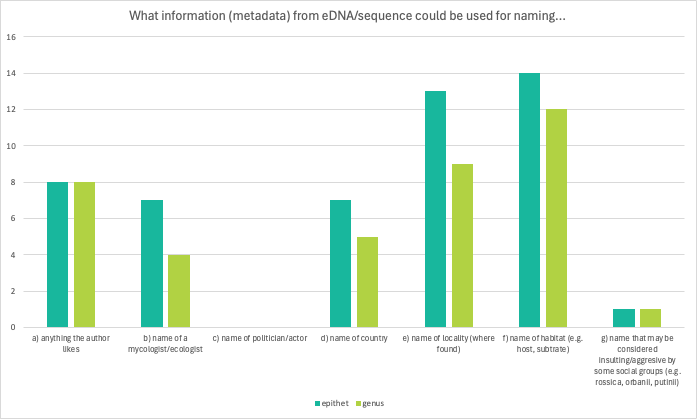
Question 4 and 5 were both about naming, the first one about epithet and second about genus. Most respondents agreed that name of locality and/or habitat could be used for both naming the epithet and the genus. None of the respondents thought it appropriate to name a new epithet or genus after a politician or actor. While some agreed that name that may be considered insulting/aggresive by some social groups could be used for naming the epithet/genus, some respondents showed their disagreement with these options by crossing them out.
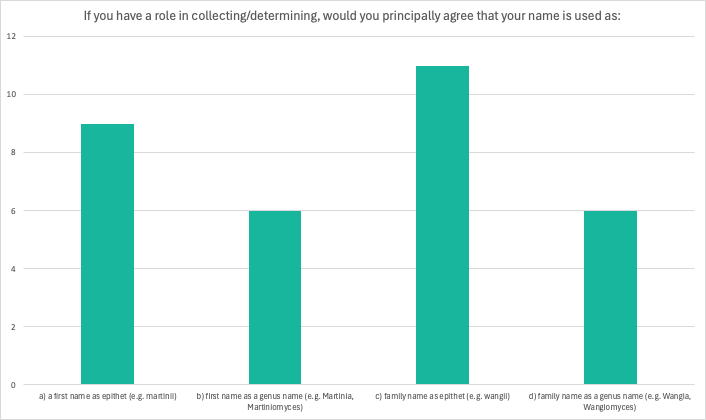
In answering question 6 most respondents agreed with using their name as an epithet or genus name, but there were also cases where some or all of the options were crossed out.
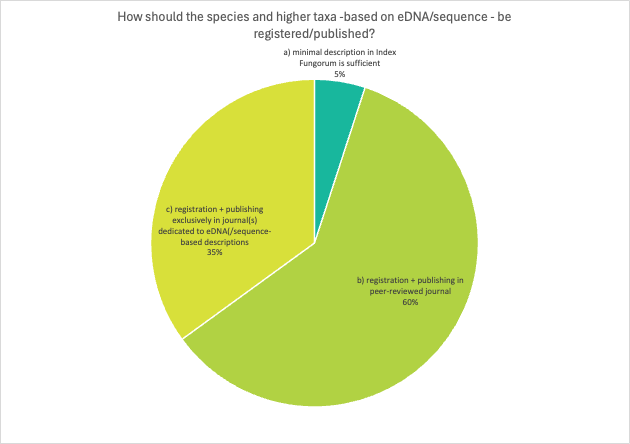
Question 7 was remarkable in a way that none of the respondents wanted to use more than one option and there is clear dominance for option b where respondents state that species and higher taxa-based on eDNA/sequence should be registered/published in peer-reviewed journals.